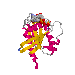 The Structural Basis for Oncogenesis
The Structural Basis for Oncogenesis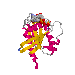 The Structural Basis for Oncogenesis
The Structural Basis for Oncogenesis
The "classic" oncogenic mutation in ras is a glycine to valine change at position 12 (gly-12 --> val) in the sequence. This mutation causes a large decrease in the GTPase activity of the ras protein. Mutations at glutamine 61 (gln-61) also decrease GTPase activity.
Mutations at a number of positions, including phe-28, asn-116, lys-117, cys-118, asp-119, thr-144 and ala-146, all cause decreases in nucleotide affinity.
It is easy to explain these observations by examining the location of these amino acid residues in the structure:
| Mutations affecting GTPase activity: | |
|
|
| Mutations affecting nucleotide affinity: | |
|
|
| Note: Only the Ca's of the ras protein are shown. The GTP is shown as a spacefilled model in cpk colors |
Notice how the residues whose mutation affects GTPase activity (gly-12 and gln-61) cluster around the g phosphate of the GTP (i.e. the area where hydrolysis occurs). It is easy to imagine that changes to the protein in these residues will distort the protein, thus moving catalytic groups out of position and decreasing the rate of the hydrolysis reaction.
Similarly, the residues whose mutation alters nucleotide binding cluster around (and interact with) the guanosine ring of the nucleotide. Again, it is easy to imagine that changes to these residues resulting in a change to this "pocket" will alter the way in which the nucleotides bind.
| Copyright © 2000, 2007 F.R. Gorga |
Last update: |